The human proteome and much more: A.I. is already revolutionizing the world
Life, from complex organisms down to their molecules, abide by the rules of mathematics and physics. This connection between fields has been explicitly recognized in science for at least 100 years, since the publishing of On Growth and Form in 1917, by the Scottish biologist and polymath D’Arcy Wentworth Thompson. In his book, Thompson explored how the laws of physics and mathematics determine the structure of organisms. Other great scientists, like Albert Einstein and Erwin Schrödinger, also acknowledged this relationship between fields, anticipating that tackling questions in one could also advance our understanding in the others. By exploring such connections, a company called DeepMind managed to use artificial intelligence to address a scientific challenge at least 50 years old: predicting the 3D structure of proteins.
Proteins are very complex molecules, and although it is possible to determine their structures experimentally, it is a time-consuming and painstaking process. Alternatively, creating computational predictions of protein structures is also challenging because the number of potential conformations of a protein grows exponentially with the length of the amino acid chain. A sequence of 100 amino acids, for example, has on the order of 1047 conformations. The development of quantum algorithms may be a key to that. However, A.I. systems are also proving useful to tackle the protein folding problem.
DeepMind has just announced the completion the free public release of the predicted structures of over 200 million proteins, a vast increase from the data on 1 million folds that it released a year ago.
DeepMind announced their new A.I. system, AlphaFold 2, in December 2020, which can use the amino-acid sequence of a protein to predict its 3D structure with atomic accuracy in just minutes. The system results from five years of development in the multidisciplinary AlphaFold team and a partnership with EMBL’s European Bioinformatics Institute. AlphaFold 2 uses deep neural networks to predict protein structures from their genetic sequence. The protein structures result from predictions of the distances between pairs of amino acids and the angles between chemical bonds that connect those amino acids. One network generates a predicted distribution of distances between every pair of residues in a protein and combines the probabilities into an estimate of the accuracy of the proposed structure. A second network then uses those distances to estimate how close the prediction is to the actual protein structure.
Using their system, DeepMind was able to publish an open-access dataset containing over 350,000 proteins. The dataset includes all ~20,000 proteins expressed by the human genome (a.k.a. the human proteome) and the proteomes of 20 other biologically significant organisms, from E. coli to mice. Additionally, they published the source code and two articles in Nature detailing how the system works.
Highly accurate protein structure prediction for the human proteome
Highly accurate protein structure prediction with AlphaFold
AlphaFold Protein Structure Database
Similar to how the structure of a machine can inform what it does, the structure of a protein help scientists understand its function. Determining the shape of proteins through experiments is extremely hard. After decades of work, we only managed to determine the structure of 17% of the proteins of the human body in the laboratory. In contrast, AlphaFold estimates that at least 36% of its predicted protein structures are accurate to the atomic level. Thus, the system effectively more than doubled our previous knowledge.
Even less accurate predictions may still be valuable. Over half of AlphaFold’s predictions for proteins of the human body should be good enough for scientists to figure out their functions. Dr. Mohammed AlQuraish, a systems biologist at Columbia University, explained that knowing the shapes of most proteins in the human body enables scientists to investigate how these proteins work as a system, not just in isolation. Additionally, the complete proteome of any newly discovered pathogen can now be available for scientists virtually immediately. Such speed provides unprecedented advantages to understanding how pathogens operate and the development of strategies to combat them.
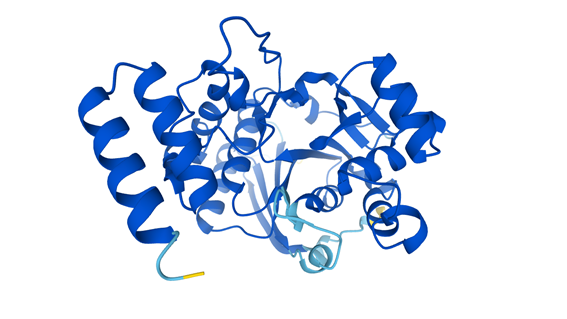
3D structure of the protein Lipopolysaccharide 1,3-galactosyltransferase. This protein is involved in the pathway that is part of the outer membrane biogenesis in the bacteria Escherichia coli.
In 2003, the publishing of the human genome impacted the world. The human genome dataset provided detailed information about the structure, organization, and function of the complete set of human genes. The human genome revolutionized our understanding of human genetics and significantly boosted researches in many fields related to human health and development. Dr. Ewan Birney, EMBL-EBI Director, declared that the human proteome is one of the most relevant datasets since the human genome. It already advanced decades-worth of work in the knowledge of the human proteome, and it carries the potential to help scientists understand diseases better and develop new treatments.
As scientists explore the structures in DeepMind’s just-announced database of over 200 million proteins , we can begin to wonder at all the possible future applications of the new knowledge. Beyond treatments for diseases and other human health-related problems, the contributions of AlphaFold have the potential to boost the development of any protein-mediated technologies, which include industrial processes, as well as problems of global importance, like microplastic pollution. Scientists worldwide are already using the dataset to advance their researches in many fields. For example, developing cures for diseases that disproportionately affect the poorer parts of the world, like Malaria, engineering proteins to recycle single-use plastics, and understanding the biology of pathogens, like the SARS-CoV-2. DeepMind expects that this almanac of proteins will revolutionize structural bioinformatics. So far, the scientists’ responses and the preliminary results derived from the datasets released earlier make us look forward to what the future holds.

3D structure of the T-cell immunomodulatory protein homolog. This protein may protect the parasite causing Malaria, Plasmodium falciparum, against attack by the host immune system.



